Priority radiomic parameters for computed tomography of head and neck malignancies: A systematic review
- Authors: Vasilev Y.A.1, Nanova O.G.1, Blokhin I.A.1, Reshetnikov R.V.1, Vladzymyrskyy A.V.1, Omelyanskaya O.V.1
-
Affiliations:
- Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
- Issue: Vol 5, No 2 (2024)
- Pages: 255-268
- Section: Systematic reviews
- Submitted: 13.11.2023
- Accepted: 04.12.2023
- Published: 20.09.2024
- URL: https://jdigitaldiagnostics.com/DD/article/view/623240
- DOI: https://doi.org/10.17816/DD623240
- ID: 623240
Cite item
Abstract
BACKGROUND: Radiomics is the newest and most promising direction in modern radiographic diagnostics. The number of head and neck cancer studies employing radiomics is increasing annually. A systematic review of recent publications (2021–2023) on computed tomography (CT) of head and neck malignancies was performed.
AIM: To present systematized data on parameters for radiomic analysis for head and neck malignancies identified by CT data.
MATERIALS AND METHODS: The literature search was carried out in PubMed. The basic characteristics of the selected articles were extracted, and their quality was assessed using RQS 2.0 and the modified QUADAS-CAD questionnaire. The reproducibility level of radiomic parameters selected for predictive models in different studies was assessed. Eleven articles were selected for the review. In most cases, a high risk of systematic error associated with data imbalance in terms of demographic parameters and level of pathologies was noted.
RESULTS: The range of RQS 2.0 scores for the included articles varied from 19.44% to 50.00% of the maximum possible score. The decreasing research quality was mainly caused by the lack of external result validation (73% of the analyzed articles) and data accessibility and transparency (82%). Inter-study reproducibility of radiomic parameters was low owing to the wide variety of techniques used for image acquisition, image post-processing, extraction, and statistical processing of radiomic parameters.
CONCLUSION: A set of stable radiomic parameters must be successfully introduced into clinical practice. The standardization of radiomics method and creation of an open radiomics database are necessary for this purpose.
Keywords
Full Text
Background
Radiomics is the latest modern medicine innovation. This technique aims to improve the diagnostic quality using radiomic features, i.e., medical image parameters invisible to the human eye [1]. Radiomics analysis is a rapidly developing area in radiology [2]. This approach is expected to be widely used as an additional tool for assessing the prognosis and determining the treatment strategy.
Several thousand radiomic parameters [3] are currently classified into three major groups:
- Curve parameters describing image properties
- Texture parameters (gray-scale matrices) representative of pixel ratios
- Shape parameters
Several subgroups were observed within each group of radiomics features.
Dozens to thousands of parameters were used in studies, depending on whether radiomic features are extracted manually or via machine learning algorithms [3, 4]. Furthermore, approaches to assigning specific parameters to major groups vary. In some cases, all groups of features can be included in varying proportions, whereas only texture parameters are included in others, excluding shape parameters. Currently, the number and composition of radiomic features during manual extraction (handcrafted features) are primarily determined using the selected analysis software and per the researcher’s perception.
The set of radiomic features should be standardized for the potential use of radiomics as an additional diagnostic tool in clinical practice [5, 6]. Features selected for a wide practical use should ensure inter-study reproducibility. However, these studies differ in various ways, including the structures examined, the type of prognosis, the method of obtaining and processing images, and the statistical analysis methods of radiomic features.
STUDY AIM
To organize data on the used radiomic parameters in head and neck cancer detected based on computed tomography (CT) findings. Head and neck cancer, including throat, larynx, nasal cavity, paranasal sinus, and oral cavity malignancies [7], was selected as one of the most common cancers [8] requiring multimodal diagnostics, beginning with CT [9–11].
Study objectives
The study objectives are as follows:
- To review the most recent publications (2021–2023) on radiomics in head and neck cancer using CT findings, including an assessment of distribution by study objectives, methods used, and article quality based on modern radiomics standards.
- To assess the intra- and inter-study reproducibility (robustness) of radiomic features.
- To compare the most recent publications with previous studies.
Materials and methods
Search Strategy
The search was performed in PubMed. The search terms were in English only. The search period—November 15, 2020, to June 1, 2023—was selected so that the reference lists of our and other studies would not overlap for the most part [12–14].
The search terms included the following:
“head and neck neoplasms” [MeSH Terms] AND (“artificial intelligence” [MeSH Terms] OR (“artificial” [All Fields] AND “intelligence” [All Fields]) OR “artificial intelligence” [All Fields] OR (“deep learning” [MeSH Terms] OR (“deep” [All Fields] AND “learning” [All Fields]) OR “deep learning” [All Fields]) OR (“machine learning” [MeSH Terms] OR (“machine” [All Fields] AND “learning” [All Fields]) OR “machine learning” [All Fields]) OR (“neural networks, computer” [MeSH Terms] OR (“neural” [All Fields] AND “networks” [All Fields] AND “computer” [All Fields]) OR “computer neural networks” [All Fields] OR (“neural” [All Fields] AND “network” [All Fields]) OR “neural network” [All Fields]) OR (“radiomic*” [All Fields]) OR “radiomic features*” [All Fields]) OR (“radiomics features*” [All Fields]) AND (“node*” [All Fields] OR “lymph node*” [All Fields] OR (“nodal” [All Fields] OR “nodally” [All Fields] OR “nodals” [All Fields]) OR “metastas*” [All Fields])
Inclusion criteria: Original research articles
Exclusion criteria: Reviews, meta-analyses, and case reports on radiomics in head and neck cancer
The study design adheres to the Preferred Reporting Items for Systematic reviews and Meta-Analyses [15].
Two experts independently reviewed the article titles and abstracts found using the search terms. This review identified several articles for full-text analysis. The third expert made the final decision in case of disagreement over including an article in the analysis. Further review of reference lists of included articles to identify eligible publications (snowballing) was not performed.
Data Extraction and Article Quality Assessment
The following information was extracted from the selected full-text articles:
- Original author and corresponding author
- Article title, year of publication, and DOI
- Journal and impact factor
- Country where the study was performed
- Study objectives
- Study design (prospective/retrospective, single-center/multicenter)
- Inclusion/exclusion criteria
- Number, sex, and age of patients
- Tumor site and type
- Total number of extracted radiomic features
- Assignment of radiomic features to classes (assessed or not assessed); if assessed, the following classes were analyzed:
– Shape parameters (2D and 3D)
– Type 1 parameters
– Type 2 parameters: texture parameters with several subgroups (Gray Level Co-occurrence Matrix [GLCM], Gray Level Run Length Matrix [GLRLM], Gray Level Size Zone Matrix [GLSZM], Neighboring Gray Tone Difference Matrix [NGTDM], and Gray Level Dependence Matrix [GLDM])
- Radiomic feature analysis method:
– Machine learning (used or not used)
– For handcrafted radiomics, statistical methods were used for the selection of radiomic features
- Number of radiomic features selected by the authors as prognostically valuable and their significance.
Two approaches were used to assess the quality of selected articles: the Radiomics Quality Score 2.0 (RQS 2.0) [16], specific to radiomics studies, and Quality Assessment of Diagnostic Accuracy Studies 2 (QUADAS-2) [17, 18], commonly used in medical studies and has been modified for computer aided detection (QUADAS-CAD).
Analysis of Radiomic Features
Radiomic features identified with prognostic value were extracted from each selected article. Features extracted from both original and post-processed images were analyzed. Various statistical methods, including machine learning, regression analysis, analysis of variance, resampling, and assessment by intraclass correlation coefficient, were used to select features to be considered. If several hypotheses were evaluated in a study, radiomic features were extracted separately for each hypothesis. Two studies provided statistics for the robustness of all extracted radiomic parameters (the intraclass correlation coefficient [19] and the p-level for the analysis of variance [20]), without reducing the number of parameters. In such cases, the most robust radiomic parameters were independently selected for our analysis, based on the available data.
Moreover, the inter- and intra-study overlap of significant radiomic feature sets was assessed for different endpoints.
Results
Literature Search and Selection of Articles
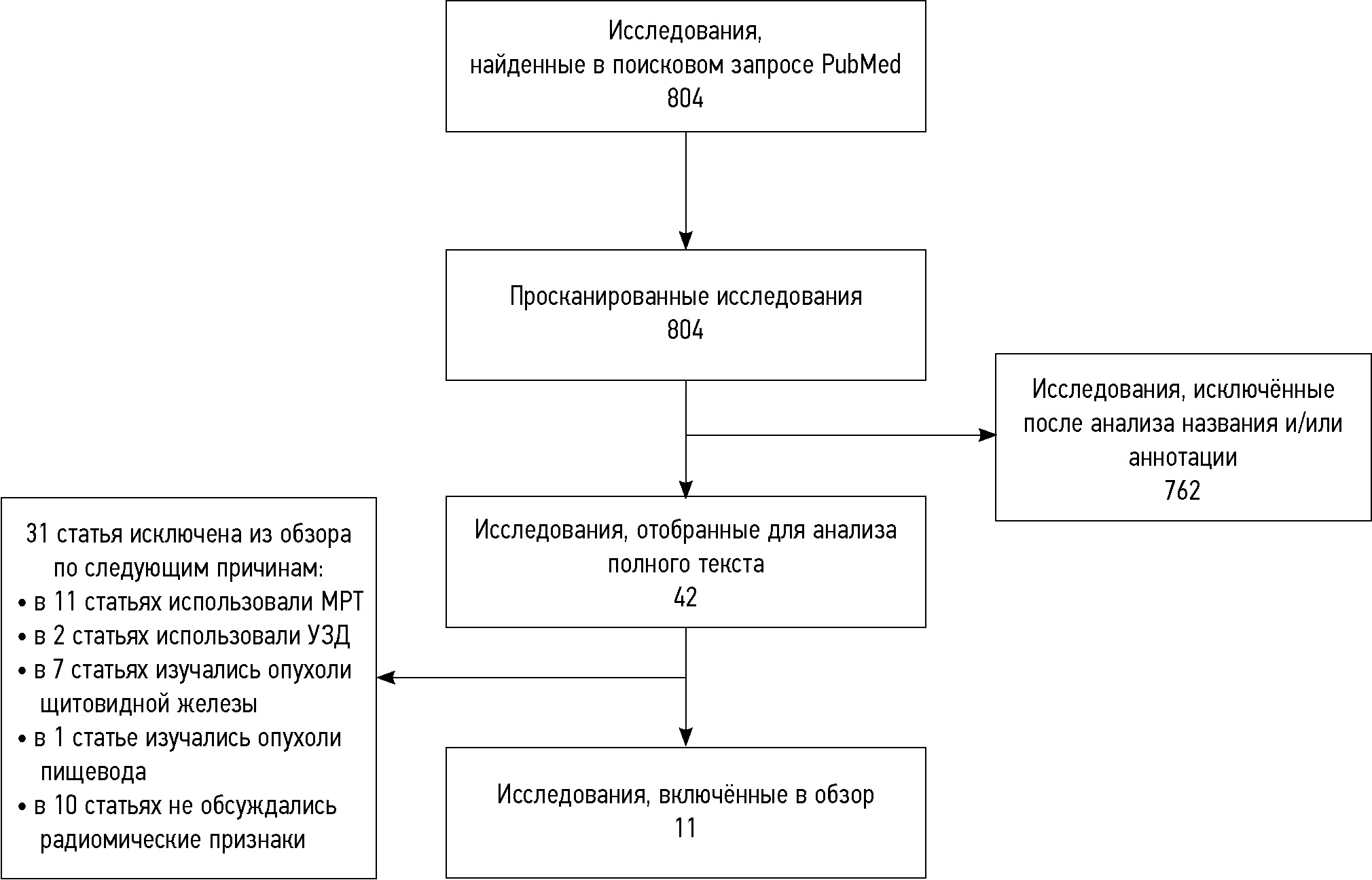
The initial search identified 804 publications. After reviewing the titles and abstracts, 762 publications were excluded as irrelevant (other types of cancer were investigated, radiomics analysis was not used, etc.). Forty-two publications were included for analysis after reviewing the titles and abstracts (Fig. 1). Of these, 11 studies were included in the final analysis, whereas 31 were excluded (11 studies used magnetic resonance imaging, 2 used ultrasound examination, 7 focused on thyroid cancer, 1 focused on esophageal cancer, and 10 did not specify the radiomic parameters used).
Fig. 1. Flow chart of systematic literature search flow chart. Abbreviations: MRI, magnetic resonance imaging; US, ultrasound.
Basic Characteristics of Articles
The basic characteristics of articles selected for the review are summarized in Appendix 1. Of these 11 studies, five were performed in China [19, 21–24], three in Europe (of them one in Italy) [25], and one each in Portugal/Austria/Germany [26], the Netherlands [27], the USA [28], Canada [20], and Thailand [29]. Articles with the highest ratings were published in Cancers (impact factor 6.575) [20] and European Radiology (impact factor 6.020) [21]. All studies were retrospective. Eight were single-center [20–23, 25, 27–29] and three were multicenter [19, 24, 26] studies.
Radiomic features were used to predict overall survival [25, 26, 29], progression-free survival [25, 29], distant disease-free survival [28], risk of locoregional recurrence [25, 26, 28], and risk of distant metastases [26]; to preoperatively predict lymph node involvement [21, 23, 24]; and to classify enlarged cervical lymph nodes [22]. One study examined the relationship between the robustness of radiomics parameters and the quality of radiomics models [18]. Another investigated the differences in radiomic features depending on the tumor site [20]. One used additional data to validate a previously created model [27].
Quality of Included Articles According to RQS 2.0
The quality assessment of articles using the specialized radiomics analysis scoring system RQS 2.0 is summarized in Appendix 2. The scores for the reviewed articles range from 7 (19.44%) [20] to 18 (50.00%) [22], with a maximum score of 36 (100%) points; the mean and standard deviation are 10 and 4, respectively.
In 7 (64%) of 11 cases, the imaging protocol was well-documented [21–26, 28]. Five studies (45%) considered the effects of segmentation (resegmentation by two researchers, segmentation algorithms, random noise) on the extraction of radiomic features [22, 23, 25, 29, 30]. Teng et al. [20] assessed the reliability of radiomic features in multicenter studies and the effects of various features on the overall reliability of models. None of the reviewed studies evaluated the robustness of radiomic features against temporal variations, such as organ movement or increased/decreased organ size. Ten (91%) studies examined model retraining and reduced number of radiomic features to select the most significant ones [19, 21–29]. Eight (73%) studies reported that models were developed using pooled sets of radiomic and clinical features and compared mixed, radiomic, and clinical models [22, 24–30]. All studies (100%) provided significance and discrimination quality metrics (area under the curve [AUC] and p-level, including those obtained during data resampling) [31]. The obtained radiomic models were rarely validated. Specifically, validation was performed in as few as three studies (27%) [20, 23, 26], with only one (9%) using data from another study site [26]. Data transparency was also limited, with only two studies providing open access to images [25] and extracted radiomic features [18].
Quality of Included Articles Based on QUADAS-CAD
The risk of bias according to QUADAS-CAD is summarized in Tables 3 and 4 [17]. The overall risk of bias was high in 6 of 11 reviewed articles (54.5%) [19, 20, 25, 26, 28, 29]. Five (45.5%) of 11 articles had a low risk of bias [21–24, 27]. Seven (64%) studies reported a high risk of bias due to data imbalance [19, 20, 23, 25, 26, 28, 29], and four (36%) reported low [21, 22, 24, 27]. In most cases, this risk was caused by a sample imbalance in terms of demographics and pathology. Machine learning was used in six studies [19, 20, 22, 24, 26, 28]; thus, some questions in D2 block are only applicable to them. The risk of bias due to the selected method for the use and interpretation of index tests was high in four studies (36%) [19, 20, 26, 29], moderate in one (9.5%) [28], and low in six (54.5%) [21–25, 27]. The risk of bias from the reference standard assessment was low in most cases (64%) [19, 21–24, 27, 29]. In some cases, the expertise level of physicians assessing the reference values was unclear; therefore, the risk of bias was considered high (27%) [28] or moderate (9%) [20, 25, 26]. The risk of bias due to data heterogeneity was high in three studies (27%) [20, 25, 28] and low in eight (73%) [19, 21–24, 26, 27, 29]. In some cases, ambiguous assessment results were due to a detailed level when describing data analysis methods.
Table 1. Quality Assessment of Diagnostic Accuracy Studies for Computer Aided Detection
Domain | Questions | Franzese С., 2023 | Gonçalves M., 2022 | Zhao X., 2023 | Teng X., 2022 | Zhang W., 2022 | Yang G., 2022 | Intarak S., 2022 | Morgan H., 2021 | Li J., 2021 | Liu X., 2021 | Zhai T., 2021 |
D1 | Were the data (training and test sets) balanced in terms of the target pathology severity (including its absence)? | No | No | Yes | Unclear | Yes | Yes | No | No | Yes | No | Yes |
Were the data (training and test sets) balanced in terms of demographic factors? | No | No | Yes | Unclear | Yes | No | No | Unclear | Yes | No | Yes | |
Was the study free of needless exclusions? | Yes | No | Yes | Unclear | Yes | Yes | Unclear | Unclear | Yes | Unclear | Yes | |
D2 | If a neural network was used, were the training and test data sets distinct or similar? | X | Yes | X | Unclear | Yes | X | X | Yes | Yes | Unclear | X |
If a neural network was used, was the size of each data set appropriate? | X | No | X | Yes | Yes | X | X | Yes | Yes | Yes | X | |
If a pathology threshold was used, was it predefined? | Yes | X | Yes | X | Yes | Yes | No | Unclear | Yes | Unclear | Yes | |
If a decision threshold was used (for AI), was it predefined? | X | X | X | X | X | X | X | Unclear | X | X | X | |
D3 | Can the reference standard accurately classify the target condition? | Unclear | Unclear | Yes | Yes | Yes | Yes | Yes | Unclear | Yes | Yes | Yes |
Were the results for reference standards generated or validated with the required expertise level? | Unclear | Unclear | Yes | Yes | Unclear | Yes | Yes | Unclear | Yes | Unclear | Yes | |
D4 | Were the results obtained in a transparent manner? | Unclear | Yes | Yes | Yes | Yes | Yes | Yes | No | Yes | Yes | Yes |
Was the same reference standard used for all patient data? | Unclear | Yes | Yes | Yes | Unclear | Yes | Yes | No | Unclear | No | Yes |
Table 4. Risk-of-bias assessment according to Quality Assessment of Diagnostic Accuracy Studies for Computer Aided Detection
Original author, year | D1 | D2 | D3 | D4 | Total score | Weight (%) |
Franzese С., 2023 | High | Low | Somewhat doubtful | High | High | 2 |
Gonçalves M., 2022 | High | High | Somewhat doubtful | Low | High | 4 |
Zhao X., 2023 | Low | Low | Low | Low | Low | 10 |
Teng X., 2022 | High | High | Low | Low | High | 32 |
Zhang W., 2022 | Low | Low | Low | Low | Low | 6 |
Yang G., 2022 | High | Low | Low | Low | Low | 4 |
Intarak S., 2022 | High | High | Low | Low | High | 4 |
Morgan H., 2021 | High | Somewhat doubtful | High | High | High | 1 |
Li J., 2021 | Low | Low | Low | Low | Low | 15 |
Liu X., 2021 | High | High | Somewhat doubtful | High | High | 14 |
Zhai T., 2021 | Low | Low | Low | Low | Low | 6 |
Methods Used in the Studies
The number of extracted radiomic features ranges from 36 [20] to 5,486 [19]. Five articles included detailed information on the distribution of extracted radiomic features [22, 23, 25, 26, 28].
Six studies used machine learning for radiomics analysis [19, 20, 22, 24, 26, 28]. The remaining five studies used regression analysis [25, 29], analysis of variance (ANOVA) [23], intraclass correlation coefficient (ICC) [29], data resampling [28], and one-way tests for pairwise comparison of features (Student’s t-test, Mann–Whitney U test, chi-squared test, Fisher’s exact test) to assess the significance of radiomic features [21, 23, 27].
The number of selected features in studies ranges from 2 [25, 27] to 19 [26]. Two articles did not select the most significant features; instead, they reported the corresponding statistics for each extracted feature (ICC) [19] and the percentage of repeated features in replicates [28].
Feature Reproducibility Analysis
In 11 studies, 191 radiomic features considered valid for prognostic models were selected (see Appendix 1), including 47 first-order features. Of these, the same feature is used in two different studies with five (11%) cases; in the remaining cases, features do not overlap between studies. Shape parameters include 25 radiomic features. Of these, the same feature is used in two different studies with five (20%) cases. Moreover, the same feature is used in three different studies in two (8%) cases. In the remaining cases, features do not overlap between studies. Second-order features include 119 radiomic features. Of these, the same feature is used in two different studies in one case (0.8%).
In two studies, radiomic features were completely reproducible between different models [23, 29]. In two more studies, radiomic features were not reproducible between different models [25, 28].
Discussion
This review analyzed studies on radiomics analysis in head and neck malignancies based on CT findings performed between 2021 and 2023, focusing on a list of frequently used, reliable radiomic parameters. The reviewed publications used a wide range of approaches, from image acquisition and post-processing methods to software used for radiomic parameter extraction and statistical processing. Furthermore, creating a predictive radiomics model always requires reducing the number of radiomic parameters. Parameters are selected using different methods, from univariate statistical tests to machine learning; this is entirely up to the authors. The selected statistical methods for reducing the number of features also have a significant impact on the selection results of parameters. The most recent meta-analyses highlighted that the difficulty of summarizing and implementing individual successful practices remains a significant barrier in current radiomics [30].
Study Quality
When comparing previous systematic reviews of radiomic studies in head and neck malignancies [13, 32] and our new study, several methodological challenges persisting for a decade were encountered.
One of the major challenges in radiomic studies is the lack of validation of obtained radiomic models using external data. Only one of the reviewed studies validated data from another study site [33].
Another major challenge is the lack of data transparency and insufficient detailed description of analysis methods, preventing the reproduction of results of such studies. However, reproducibility of results is widely considered one of the fundamental criteria of scientific approach and the basis for practical implementation of a method [34].
Our conclusions are consistent with the findings of other systematic reviews. For example, all four identified reviews of studies evaluating head and neck cancer [12, 13, 32, 35] lacked result validation using external data. Moreover, Giannitto et al. [13] reported a lack of transparency of methodologies used in studies due to an inadequate description of the study conduct and a lack of assessment of the possible implementation of results in clinical practice. Guha et al. [12] revealed substantial heterogeneity of methodologies, making it difficult to generalize study findings.
The Image Biomarker Standardization Initiative (IBSI) is currently in progress [36]. Considering the detailed level in addressing the issues and numbers of community members involved, this initiative could be a significant step toward resolving the lack of transparency in radiomic analysis. The reproducibility and reliability of results can also be improved by appropriately designed clinical studies of intelligent technology-based algorithms [37].
Creating an open platform for radiomic studies will enable reporting negative results, which are not published in peer-reviewed journals due to the so-called publication bias [38]. Minimizing the risk of bias is critically important when assessing the efficacy of radiomic analysis. Furthermore, Kocak et al.’s [39] meta-study made it possible to highlight issues caused by a largely retrospective design (95%, 142/149) and a lack of a reference test in a significant number of studies (44%, 66/149) [39].
The assessed method is based on radiomic parameters describing the relationships among voxels, 2D and 3D characteristics of malignancies, and other properties. Several thousands of these parameters are currently known; however, the consensus regarding the diagnostic value of each parameter and its various combinations is not yet established. The number of selected features in reviewed studies dramatically varies, ranging from a few to several thousands. Less than half of studies provide detailed descriptions of the groups of features representing various characteristics of malignancies. Three studies did not specify the radiomic parameters used in the models. Only one reviewed article examined the robustness of radiomic parameters in multicenter studies.
To promote the widespread use of prognostic radiomics models in clinical practice, priority parameters must be identified based on their robustness and reproducibility assessment. Radiomic parameters most used in prognostic models were selected. Our findings demonstrated that the reproducibility of radiomic parameters is extremely low due to the wide range of methodologies used. This is consistent with earlier studies suggesting that radiomic parameters might be random and non-reproducible [40]. Recommending a specific set of radiomic parameters for clinical use is difficult. Therefore, radiomic methods must be standardized, and recommended standards must be implemented. Consequently, a basic set of radiomic parameters can be created for the use of radiomic analysis in diagnostic imaging [41]. Standardization of radiomic analysis can also be achieved through efforts in the field of study protocol standardization and post-processing control standardization [42].
Limitations of Our Approach
Our study has several limitations inherent for systematic reviews. With the aim to provide the most comprehensive review of currently available studies of head and neck malignancies, this review includes studies of both primary and secondary tumors and histologically heterogeneous head and neck cancers.
The search was limited to PubMed and English publications, which may have reduced the number of identified studies.
Data imbalance was observed in all studies. Only pathological cases were included, while non-pathological cases were excluded. Moreover, data imbalance was also observed in demographics.
These limitations prevented comprehensive meta-analysis, allowing only qualitative synthesis with descriptive statistics. However, our study highlighted the major challenges of modern radiomics and the direction of future research in this area.
Conclusion
Radiomics is a rapidly evolving modern medicine area. Studies increasingly used radiomics analysis. Our findings revealed that major challenges in this area preventing the wide clinical use of this promising method include low transparency of studies and the absence of open-access databases and standardized approaches to radiomics studies. The fundamental objective of radiomics development is to adopt accepted standards for image acquisition and processing, as well as modeling strategies. Assessment tools for the risk of bias should be used during studies, such as QUADAS-2 or its versions modified for specific tasks, and recommendations should be considered for reducing these risks. Free access to radiomics data should be enabled, similarly to genetic studies. A set of robust radiomic parameters should be developed to use this method in clinical practice. The IBSI platform is an effective solution for the standardization of radiomics data and its open-access publication.
Additional information
Additional materials.
Supplement 1. Basic characteristics of articles.
DOI: https://doi.org/10.17816/DD623240-4214843
Suplement 2. Radiomics quality assessment according to RQS-2.0. DOI: https://doi.org/10.17816/DD623240-4214842
Funding source. This paper was prepared by a group of authors as a part of the research and development effort titled “Scientific evidence for using radiomics-guided medical imaging to diagnose cancer”, No. 123031400009-1” (USIS No. 123031500005-2), in accordance with the Order No. 1196 dated December 21, 2022 “On approval of state assignments funded by means of allocations from the budget of the city of Moscow to the state budgetary (autonomous) institutions subordinate to the Moscow Health Care Department, for 2023 and the planned period of 2024 and 2025” issued by the Moscow Health Care Department.
Competing interests. The authors declare no сompeting interests.
Authors’ contribution. All authors confirm that their authorship meets the international ICMJE criteria (all authors made a significant contribution to the development of the concept, conduct of the study and preparation of the article, read and approved the final version before publication. The contribution is distributed as follows: Yu.A. Vasilev, A.V. Vladzymyrskyy, O.V. Omelyanskaya — study concept, approval of the final version of the manuscript; R.V. Reshetnikov, I.A. Blokhin, O.G. Nanova — literature review, data analysis, writing the text of the article.
About the authors
Yuriy A. Vasilev
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: npcmr@zdrav.mos.ru
ORCID iD: 0000-0002-0208-5218
SPIN-code: 4458-5608
MD, Cand. Sci. (Medicine)
Russian Federation, MoscowOlga G. Nanova
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Author for correspondence.
Email: NanovaOG@zdrav.mos.ru
ORCID iD: 0000-0001-8886-3684
SPIN-code: 6135-4872
Cand. Sci. (Biology)
Russian Federation, MoscowIvan A. Blokhin
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: i.blokhin@npcmr.ru
ORCID iD: 0000-0002-2681-9378
SPIN-code: 3306-1387
Russian Federation, Moscow
Roman V. Reshetnikov
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: reshetnikov@fbb.msu.ru
ORCID iD: 0000-0002-9661-0254
SPIN-code: 8592-0558
Cand. Sci. (Physics and Mathematics)
Russian Federation, MoscowAnton V. Vladzymyrskyy
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: npcmr@zdrav.mos.ru
ORCID iD: 0000-0002-2990-7736
SPIN-code: 3602-7120
MD, Dr. Sci. (Medicine), Professor
Russian Federation, MoscowOlga V. Omelyanskaya
Research and Practical Clinical Center for Diagnostics and Telemedicine Technologies
Email: o.omelyanskaya@npcmr.ru
ORCID iD: 0000-0002-0245-4431
SPIN-code: 8948-6152
Russian Federation, Moscow
References
- Rizzo S, Botta F, Raimondi S, et al. Radiomics: the facts and the challenges of image analysis. Eur Radiol Exp. 2018;2. doi: 10.1186/s41747-018-0068-z
- Lambin P, Leijenaar R, Deist T, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14:749–762. doi: 10.1038/nrclinonc.2017.141
- Mayerhoefer ME, Materka A, Langs G, et al. Introduction to Radiomics. J Nucl Med. 2020;61(4):488–495. doi: 10.2967/jnumed.118.222893
- Wan Y, Yang P, Xu L, et al. Radiomics analysis combining unsupervised learning and handcrafted features: A multiple-disease study. Medical Physics. 2021;48(11):7003–7015. doi: 10.1002/mp.15199
- Zwanenburg A, Vallières M, Abdalah MA, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology. 2020;295(2). doi: 10.1148/radiol.2020191145
- Berenguer R, Pastor-Juan M, Canales-Vázquez J, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology. 2018;288(2). doi: 10.1148/radiol.2018172361
- Laura QM, Chow MD. Head and Neck Cancer. N Engl J Med. 2020;382:60–72. doi: 10.1056/NEJMra1715715
- Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Ca Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660
- Bolotina LV, Vladimirova LYu, Den’gina NV, Novik AV, Romanov IS. Practice guidelines for the treatment of malignant tumors of the head and neck. Zlokačestvennye opuholi. 2020;10(3s2-1):93–108. (In Russ). doi: 10.18027/2224-5057-2020-10-3s2-06
- Bossi P, Chan AT, Licitra L, et al. Nasopharyngeal carcinoma: ESMO-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology. 2021;32(4):452–465. doi: 10.1016/j.annonc.2020.12.007
- Petrovichev VS, Neklyudova MV, Sinitsyn VE, Nikitin IG. Dual-energy computed tomography for head and neck cancer. Digital Diagnostics. 2021;2(3):343–355. doi: 10.17816/DD62572
- Guha A, Connor S, Anjari M, et al. Radiomic analysis for response assessment in advanced head and neck cancers, a distant dream or an inevitable reality? A systematic review of the current level of evidence. BJR. 2019;93(1106). doi: 10.1259/bjr.20190496
- Giannitto C, Mercante G, Ammirabile A, et al. Radiomics-based machine learning for the diagnosis of lymph node metastases in patients with head and neck cancer: Systematic review. Head & Neck. 2022;45(2):482–491. doi: 10.1002/hed.27239
- Yu Y, Li X, Li W, Wang H, Wang Y. Pretreatment radiomics power in evaluating neoadjuvant chemotherapy response and outcome for patients with head and neck squamous cell carcinoma: a systematic review and meta-analysis. Preprint. 2023. doi: 10.21203/rs.3.rs-2530190/v1
- Page MJ, McKenzie JE, Bossuyt PM, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ. 2021;372. doi: 10.1136/bmj.n71
- Radiomics Quality Score — RQS 2.0 [Internet]. [cited 1 Jan 2023]. Available from: https://www.radiomics.world/rqs2
- Kodenko MR, Vasilev YA, Vladzymyrskyy AV, et al. Diagnostic Accuracy of AI for Opportunistic Screening of Abdominal Aortic Aneurysm in CT: A Systematic Review and Narrative Synthesis. Diagnostics. 2022;12(12):3197. doi: 10.3390/diagnostics12123197
- Kodenko MR, Reshetnikov RV, Makarova TA. Modification of quality assessment tool for artificial intelligence diagnostic test accuracy studies (QUADAS-CAD). Digital Diagnostics. 2022;3(S1):4–5. doi: 10.17816/DD105567
- Teng X, Zhang J, Ma Z, et al. Improving radiomic model reliability using robust features from perturbations for head-and-neck carcinoma. Front. Oncol. 2022;12. doi: 10.3389/fonc.2022.974467
- Liu X, Maleki F, Muthukrishnan N, et al. Site-Specific Variation in Radiomic Features of Head and Neck Squamous Cell Carcinoma and Its Impact on Machine Learning Models. Cancers. 2021;13(15). doi: 10.3390/cancers13153723
- Zhao X, Li W, Zhang J, et al. Radiomics analysis of CT imaging improves preoperative prediction of cervical lymph node metastasis in laryngeal squamous cell carcinoma. European Radiology. 2023;33:1121–1131. doi: 10.1007/s00330-022-09051-4
- Zhang W, Peng J, Zhao S, et al. Deep learning combined with radiomics for the classification of enlarged cervical lymph nodes. Journal of Cancer Research and Clinical Oncology. 2022;148:2773–2780. doi: 10.1007/s00432-022-04047-5
- Yang G, Yang F, Zhang F, et al. Radiomics Profiling Identifies the Value of CT Features for the Preoperative Evaluation of Lymph Node Metastasis in Papillary Thyroid Carcinoma. Diagnostics. 2022;12(5). doi: 10.3390/diagnostics12051119
- Li J, Wu X, Mao N, et al. Computed Tomography-Based Radiomics Model to Predict Central Cervical Lymph Node Metastases in Papillary Thyroid Carcinoma: A Multicenter Study. Front. Endocrinol. 2021;12. doi: 10.3389/fendo.2021.741698
- Franzese C, Lillo S, Cozzi L, et al. Predictive value of clinical and radiomic features for radiation therapy response in patients with lymph node-positive head and neck cancer. Head & Neck. 2023;45(5):1184–1193. doi: 10.1002/hed.27332
- Gonçalves M, Gsaxner Ch, Ferreira A, et al. Radiomics in Head and Neck Cancer Outcome Predictions. Diagnostics. 2022;12(11). doi: 10.3390/diagnostics12112733
- Zhai T, Wesseling F, Langendijk J, et al. External validation of nodal failure prediction models including radiomics in head and neck cancer. Oral Oncology. 2021;112. doi: 10.1016/j.oraloncology.2020.105083
- Morgan HE, Wang K, Dohopolski M, et al. Exploratory ensemble interpretable model for predicting local failure in head and neck cancer: the additive benefit of CT and intra-treatment cone-beam computed tomography features. Quant Imaging Med Surg. 2021;11(12):4781–4796. doi: 10.21037/qims-21-274
- Intarak S, Chongpison Y, Vimolnoch M, et al. Tumor Prognostic Prediction of Nasopharyngeal Carcinoma Using CT-Based Radiomics in Non-Chinese Patients. Front. Oncol. 2022;12. doi: 10.3389/fonc.2022.775248
- Zhong J, Lu J, Zhang G, et al. An overview of meta-analyses on radiomics: more evidence is needed to support clinical translation. Insights Imaging. 2023;14. doi: 10.1186/s13244-023-01437-2
- Certificate of state registration of the computer program № 2022617324/ 19.04.2022. Morozov SP, Andreichenko AE, Chetverikov SF, et al. Web-based tool for performing ROC analysis of diagnostic test results: № 2022616046. (In Russ).
- Giraud P, Giraud Ph, Gasnier A, et al. Radiomics and Machine Learning for Radiotherapy in Head and Neck Cancers. Front. Oncol. 2019;9. doi: 10.3389/fonc.2019.00174
- Certificate of state registration of the computer program № 2023619686/ 15.05.2023. Vasilev YuA, Vladzymirskyy AV, Omelyanskaya OV, et al. Web-based tool for quality control of datasets: № 2023617136. (In Russ).
- Gundersen OE, Kjensmo S. State of the Art: Reproducibility in Artificial Intelligence. Proceedings of the AAAI Conference on Artificial Intelligence. 2018;32(1). doi: 10.1609/aaai.v32i1.11503
- Tortora M, Gemini L, Scaravilli A, et al. Radiomics Applications in Head and Neck Tumor Imaging: A Narrative Review. Cancers. 2023;15(4). doi: 10.3390/cancers15041174
- Image Biomarker Standardisation Initiative (IBSI) [Internet]. [cited 1 Jan 2023]. Available from: https://theibsi.github.io/
- Morozov SP, Vladzymirskyy AV, Klyashtornyi VG, et al. Practice guidelines for the treatment of malignant tumors of the head and neck. Moscow; 2019. (In Russ).
- Buvat I, Orlhac F. The Dark Side of Radiomics: On the Paramount Importance of Publishing Negative Results. The Journal of Nuclear Medicine. 2019;60(11):1543–1544. doi: 10.2967/jnumed.119.235325
- Kocak B, Bulut E, Bayrak ON, et al. NEgatiVE results in Radiomics research (NEVER): A meta-research study of publication bias in leading radiology journals. European Journal of Radiology. 2023;163. doi: 10.1016/j.ejrad.2023.110830
- Berenguer R, Pastor-Juan MR, Canales-Vázquez J, et al. Radiomics of CT Features May Be Nonreproducible and Redundant: Influence of CT Acquisition Parameters. Radiology. 2018;288(2). doi: 10.1148/radiol.2018172361
- Morozov SP, Lindenbraten LD, Gabai PG, et al. Fundamentals of medical imaging management. Morozov SP, editor. Moscow: GEOTAR-Media; 2020. (In Russ). doi: 10.33029/9704-5247-9-MEN-2020-1-424
- Khoruzhaya AN, Ahkmad ES, Semenov DS. The role of the quality control system for diagnostics of oncological diseases in radiomics. Digital Diagnostics. 2021;2(2):170–184. doi: 10.17816/DD60393
Supplementary files